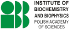Available Saccharomyces cerevisiae transcripts
4470 records found
| Gene Id | Other IDs | Gene Description↓ | |
|---|---|---|---|
| YPL020C | ULP1; SGDID:S000005941 | Ubl (ubiquitin-like protein)-specific protease that cleaves Smt3p protein conjugates; specifically required for cell cycle pr... | |
| YBR058C | UBP14; SGDID:S000000262 | Ubiquitin-specific protease that specifically disassembles unanchored ubiquitin chains; involved in fructose-1,6-bisphosphata... | |
| YOR124C | UBP2; SGDID:S000005650 | Ubiquitin-specific protease that removes ubiquitin from ubiquitinated proteins; interacts with Rsp5p and is required for MVB ... | |
| YDL122W | UBP1; SGDID:S000002280 | Ubiquitin-specific protease that removes ubiquitin from ubiquitinated proteins; cleaves at the C terminus of ubiquitin fusion... | |
| YMR304W | UBP15; SGDID:S000004920 | Ubiquitin-specific protease that may play a role in ubiquitin precursor processin; Verified ORF. | |
| YMR223W | UBP8; SGDID:S000004836 | Ubiquitin-specific protease that is a component of the SAGA (Spt-Ada-Gcn5-Acetyltransferase) acetylation complex; required fo... | |
| YER151C | UBP3; SGDID:S000000953 | Ubiquitin-specific protease that interacts with Bre5p to co-regulate anterograde and retrograde transport between endoplasmic... | |
| YNL186W | UBP10; SGDID:S000005130 | Ubiquitin-specific protease that deubiquitinates ubiquitin-protein moieties; may regulate silencing by acting on Sir4p; invol... | |
| YIL156W | UBP7; SGDID:S000001418 | Ubiquitin-specific protease that cleaves ubiquitin-protein fusion; Verified ORF. | |
| YFR010W | UBP6; SGDID:S000001906 | Ubiquitin-specific protease situated in the base subcomplex of the 26S proteasome, releases free ubiquitin from branched poly... | |
| YGR003W | CUL3; SGDID:S000003235 | Ubiquitin-protein ligase, member of the cullin family with similarity to Cdc53p and human CUL3; required for ubiquitin-depend... | |
| YDR143C | SAN1; SGDID:S000002550 | Ubiquitin-protein ligase, involved in the proteasome-dependent degradation of aberrant nuclear protein; Verified ORF. | |
| YOL013C | HRD1; SGDID:S000005373 | Ubiquitin-protein ligase required for endoplasmic reticulum-associated degradation (ERAD) of misfolded proteins; genetically ... | |
| YIL030C | SSM4; SGDID:S000001292 | Ubiquitin-protein ligase involved in ER-associated protein degradation; located in the ER/nuclear envelope; ssm4 mutation sup... | |
| YKL010C | UFD4; SGDID:S000001493 | Ubiquitin-protein ligase (E3) that interacts with Rpt4p and Rpt6p, two subunits of the 19S particle of the 26S proteasome; cy... | |
| YGR184C | UBR1; SGDID:S000003416 | Ubiquitin-protein ligase (E3) that interacts with Rad6p/Ubc2p to ubiquitinate substrates of the N-end rule pathway; binds to ... | |
| YIL008W | URM1; SGDID:S000001270 | Ubiquitin-like protein with weak sequence similarity to ubiquitin; depends on the E1-like activating enzyme Uba4p; molecular ... | |
| YDR139C | RUB1; SGDID:S000002546 | Ubiquitin-like protein with similarity to mammalian NEDD8; conjugation (neddylation) substrates include the cullins Cdc53p, R... | |
| YNR032C-A | HUB1; SGDID:S000007251 | Ubiquitin-like protein modifier, may function in modification of Sph1p and Hbt1p, functionally complemented by the human or S... | |
| YGL087C | MMS2; SGDID:S000003055 | Ubiquitin-conjugating enzyme variant involved in error-free postreplication repair; forms a heteromeric complex with Ubc13p, ... |