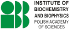Available Saccharomyces cerevisiae transcripts
4470 records found
| Gene Id | Other IDs | Gene Description | |
|---|---|---|---|
| YMR088C | VBA1; SGDID:S000004694 | Permease of basic amino acids in the vacuolar membran; Verified ORF. | |
| YMR089C | YTA12; SGDID:S000004695 | Component, with Afg3p, of the mitochondrial inner membrane m-AAA protease that mediates degradation of misfolded or unassembl... | |
| YMR090W | SGDID:S000004696 | Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; GFP-fusion protein localizes to the cy... | |
| YMR091C | NPL6; SGDID:S000004697 | Component of the RSC chromatin remodeling complex; interacts with Rsc3p, Rsc30p, Ldb7p, and Htl1p to form a module important ... | |
| YMR092C | AIP1; SGDID:S000004698 | Actin cortical patch component, interacts with the actin depolymerizing factor cofilin; required to restrict cofilin localiza... | |
| YMR093W | UTP15; SGDID:S000004699 | Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of... | |
| YMR097C | MTG1; SGDID:S000004703 | Peripheral GTPase of the mitochondrial inner membrane, essential for respiratory competence, likely functions in assembly of ... | |
| YMR098C | ATP25; SGDID:S000004704 | Mitochondrial protein required for the stability of Oli1p (Atp9p) mRNA and for the Oli1p ring formation; YMR098C is not an es... | |
| YMR099C | SGDID:S000004705 | Glucose-6-phosphate 1-epimerase (hexose-6-phosphate mutarotase), likely involved in carbohydrate metabolism; GFP-fusion prote... | |
| YMR100W | MUB1; SGDID:S000004706 | Protein that interacts with E2 and E3 enzymes Ubr2p and Rad6p and is required for ubiquitylation and degradation of Rpn4p; co... | |
| YMR102C | SGDID:S000004708 | Protein of unknown function; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes ... | |
| YMR104C | YPK2; SGDID:S000004710 | Protein kinase with similarity to serine/threonine protein kinase Ypk1p; functionally redundant with YPK1 at the genetic leve... | |
| YMR105C | PGM2; SGDID:S000004711 | Phosphoglucomutase, catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose m... | |
| YMR106C | YKU80; SGDID:S000004712 | Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position... | |
| YMR110C | HFD1; SGDID:S000004716 | Putative fatty aldehyde dehydrogenase, located in the mitochondrial outer membrane and also in lipid particles; has similarit... | |
| YMR111C | SGDID:S000004717 | Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; YMR111C is not an essen... | |
| YMR112C | MED11; SGDID:S000004718 | Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II hol... | |
| YMR113W | FOL3; SGDID:S000004719 | Dihydrofolate synthetase, involved in folic acid biosynthesis; catalyzes the conversion of dihydropteroate to dihydrofolate i... | |
| YMR114C | SGDID:S000004720 | Protein of unknown function; may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GF... | |
| YMR115W | MGR3; SGDID:S000004721 | Subunit of the mitochondrial (mt) i-AAA protease supercomplex, which degrades misfolded mitochondrial proteins; forms a subco... |